User Guides
- Chambers and Coverslips
- Fluorescent Dyes
- Fluorescent Proteins
- Immunofluorescence
Fluorescent proteins are completely genetically encoded. Therefore, cells can be studied live with minimum perturbations and labelling is 100 % specific. The University of California San Diego has a history of developing fluorescent proteins which is emphasized by the awarded Nobel Prize to Roger Tsien in 2008. Currently, there are many fluorescent proteins available, however it is very common to just grab any construct you can find in your lab’s freezers. We would like to emphasis that selecting the appropriate fluorescent protein is crucial to the quality of your microscopy data. It might seem like a lot of effort to clone new constructs, but it will be very rewarding when your microscopy analysis shows robust and reproducible numbers.
Here we discuss the most important aspects:
The FP base is a great tool to explore the variety of fluorescent proteins. Once you have selected a fluorescent protein, you can order a construct containing the sequence from Addgene or the gene from a nucleotide manufacturer (e.g. Integrated DNA Technologies). For the lasers and LEDs available in the Nikon Imaging Center, the following fluorescent proteins are a good option: mTagBFP2 (blue), mTurquoise2 (cyan), mEmerald/sGFP2/mNeonGreen (green), mVenus (yellow), mScarlet-I (red) and miRFP670 (far-red, requires biliverdin as a cofactor). Note, mTurquoise2 (cyan) and mVenus (yellow) can only be used with Cyril and Krieger. Additionally, it is possible to use photoactivatable and photoconvertible proteins such as mPA-GFP or mEos4a for protein tracking and PALM experiments.
Next, also consider the length and flexibility between the fluorescent protein and the protein you want to study. It may be useful to test several linker lengths and the positioning at the N- and C-terminus to create a tagged protein that behaves like the endogenous protein. For CRISPR strategies, it might be easier to add the short nucleotide encoding the 11th β-sheet to an endogenous locus by using a split FP variant. For low concentrations and single molecule detection, it can be beneficial to add multiple copies of the fluorescent reporter to enhance the signal. For semi-genetically encoded tags, the SNAP, CLIP, and Halo-tags can used with a variety of bright cell permeant and impermeant organic dyes, see the next section for selecting fluorescent dyes. Feel free to ask as advice specifically for your project.
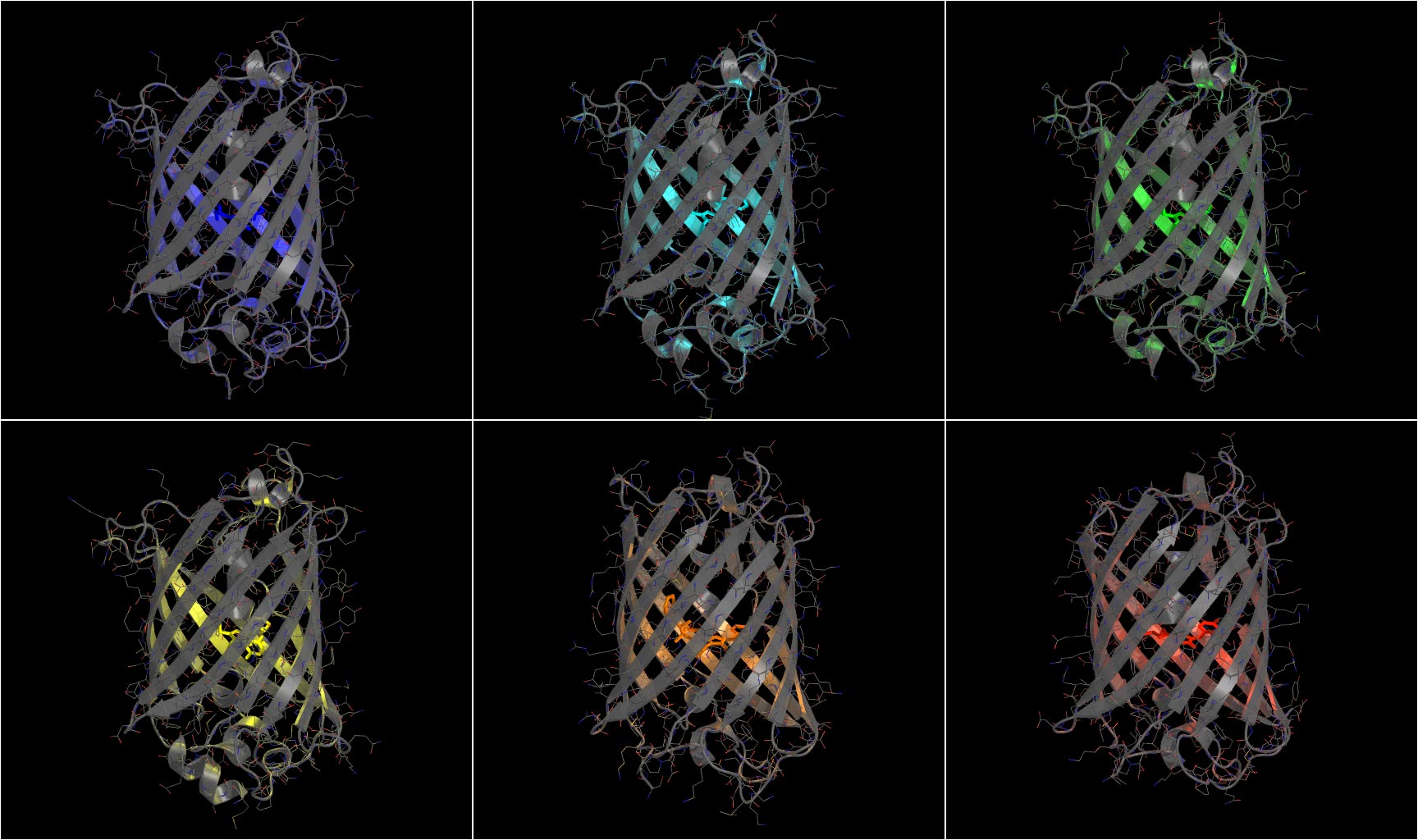
Crystal structures of fluorescent proteins by Marten Postma.